NFORM = <format>
where <format> is one of:
MDYN- MDynaMix binary trajectory (default)XMOL- XMOL trajectory. It is implied, that the commentary (second) line of each configuration is written in the format:(char) <time> (char-s) BOX: <box_x> <box_y> <box_z>
where
(char)is any character word,<time>is time in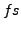 ,
,
<box_x> <box_y> <box_z>(following after keywordBOX) are the box sizes.PDBT- PDB trajectory as generated by ``trajconv'' utility of GROMACS simulation packageDCDT- DCD trajectories generated by NAMD package
FNAME = <file_name>
set the base name of the trajectory files. The trajectory must be written
as a sequence of files <file_name>.001 , <file_name>.002 and
so on, the largest possible number being <file_name>.999 .
PATHDB = <value>
Directory with molecular description files (.mmol). Default is the current directory (.) .
NTYPES = <value>
Number of molecule types in the trajectory
NAMOL = <name1> [,<name2>,...]
NTYPES names of molecules. It is supposed that files
<name1>.mmol, <name2>.mmol,... describing the molecules are
present in the directory defined by PATHDB. Format of .mmol files is
the same as for MDynaMix program. For analyzing trajectories generated by
other programs, .mmol files are still needed. It is however enough to have only
the first section of .mmol files containing names of atoms.
NSPEC = <n1>[,<n2>,...]
Number of molecules of each type ( NTYPES numbers). This parameter
is not necessary in MDynaMix binary trajectories.
NFBEG = <value>
Number of the first trajectory file (integer between 0 and 999)
NFEND = <value>
Number of the last trajectory file (integer between 0 and 999)
IPRINT = <value>
Defines how much you see in the intermediate output. The final output with analysis of results does not depend on it. Default value is 5.
BOXL = <x-box-size> BOYL = <y-box-size> BOZL = <z-box-size>define the box size if it is not present in the trajectory (implies constant-volume simulation)
BREAKM = <value>
Defines which break in trajectory (in ps) is allowed to consider the trajectory as continuous. The parameter is used in analysis of time-dependent properties.
LVEL = <logical>
Accept a logical value. .true. signals that the trajectory
may contain velocities, which will then be read for the analysis.
LXMOL = <logical>
Accept a logical value. If .true., rewrite trajectory in XMOL format
FXMOL = <name>
File name for output XMOL trajectory
ISTEP = <value>
Specifies that only each ISTEP-th configuration from the trajectory
is taken for the analysis
LBOND = <logical>
Accept logical value. If .true., information about bonds is read from .mmol files
VFAC = <value>
Multiply velocities from the trajectory by the given <value>,
in order to bring them to Å/fs (the units which are used in the analysis).
Default is 1.