Protein folding on PC
Although the problem of protein folding is far from being solved in generalterms, this process can be simulated for simple stable proteins.It is exemplified by small alpha (tryptophan cage) protein. Trp-Cage was shown to fold in several µs in experiment.Explicit water simulations give similar results. Unfortunately, such simulations require huge calculation time. The calculation time can be reduced by several orders of magnitude by using implicit water models.This interferes with the calculation accuracy, but simulation can be performed on a personal computer.
Abalone supports several force fields applicable to numerical experiments on protein folding. The table below shows the force fields thatallowed us to fold some proteins into their native conformations.
Examples of successfully folded proteins:
| Force Field | Trp-Cage α-protein | Chignolin β-protein | 2I9M α-protein |
| AMBER-ii | + | + | + |
| AMBER99SB | + | + | |
| AMBER03 | + | 4 ns, 8 replica | |
| AMBER94 | + | ||
| AMBER96 | 1.5 ns, 8 replica | ||
| OPLS | +- | ||
| OPLS-H | +- |
Trp-Cage NLYIQWLKDGGPSSGRPPPS
Chignolin GYDPETGTWG
2I9M SAAEAYAKRIAEAMAKG
Trp-Cage
First, a protein model should be generated. Open the Chain Builder(Build > Chain menu). Select Amino Acids. Input the Trp-Cage sequence (NLYIQWLKDGGPSSGRPPPS) and press the Apply button.
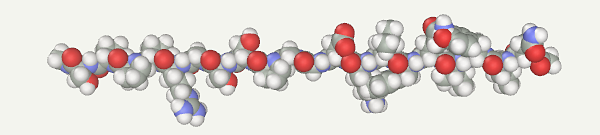
Select the AMBER94 force field (Settings > Force field menu) and make sure that the implicit water model is enabled (Settings > Implicit water menu; check box Implicit water and combo box Generalized Born).
Atoms of amino acid side radicals can overlap during the assembly. Such overlapping can be obviated by a preliminary 20-step optimization.Start optimization by clicking remove clashes  button.Apart form new models, short optimization is recommended after the force field is changed, since the model can appear in a high-energy state in terms ofanother force field. Accordingly, the force field was specified before the optimization.
button.Apart form new models, short optimization is recommended after the force field is changed, since the model can appear in a high-energy state in terms ofanother force field. Accordingly, the force field was specified before the optimization.
After the initial model was generated, a molecular dynamics simulation can be started. Invoke the Compute > Dynamics panel. Set the temperatureto 350 K and simulation time to 10 000 ps (10 ns), and start the process by Run button .Commonly, 3-10 ns suffice to reach the native protein conformation. The simulation lasts a day on a typical personal computer.
.Commonly, 3-10 ns suffice to reach the native protein conformation. The simulation lasts a day on a typical personal computer.
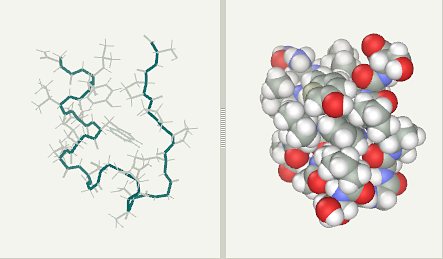
Trp-Cage experimental (pdb1L2Y NMR)
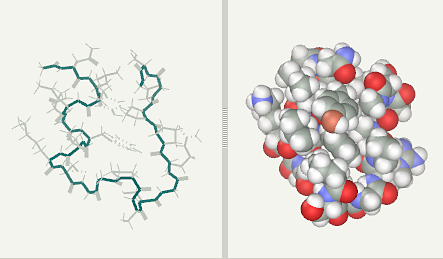
Trp-Cage folding simulation after 2 ns (file)
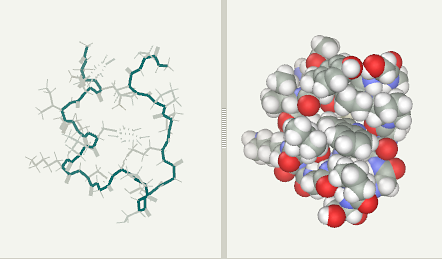
Trp-Cage folding simulation after 4 ns(file)
 Copyright © 2006-2011 Agile Molecule. All rights reserved
Copyright © 2006-2011 Agile Molecule. All rights reserved